TWO MAJOR APPROACHES
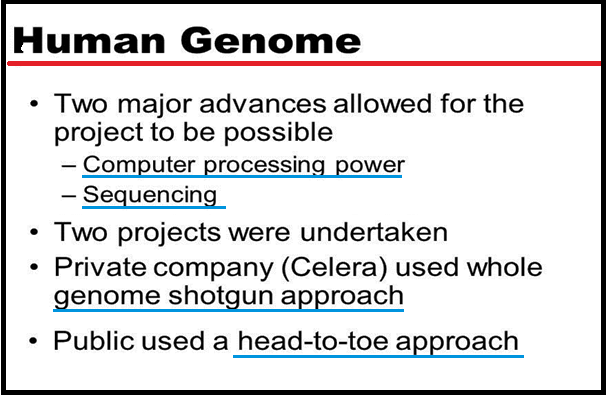
● The methods involved in HGP used `color{Violet}"two major approaches"`.
● One approach focused on identifying all the `color{Violet}"genes"` that expressed as `color{Violet}"RNA"` (referred to as `color{Violet}"Expressed Sequence Tags"` (`color{Violet}"ESTs"`).
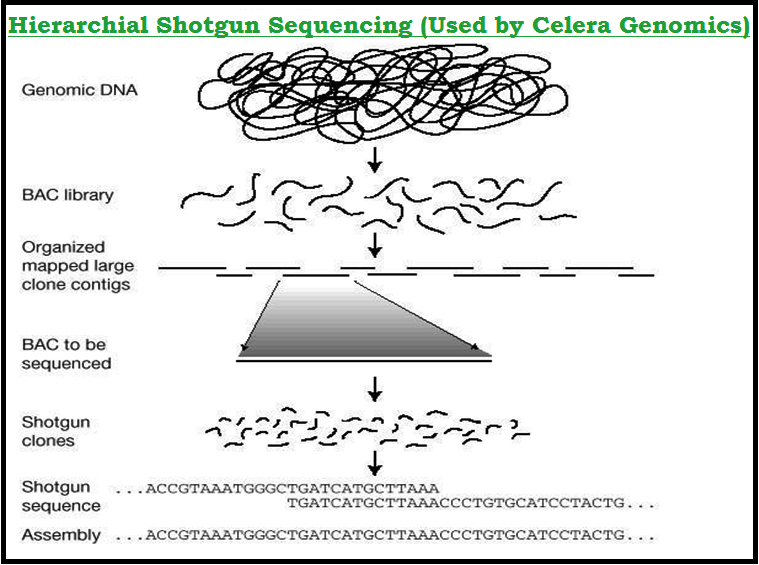
● The other took the `color{Violet}"blind approach"` of simply sequencing the `color{Violet}"whole set of genome"` that contained all the `color{Violet}"coding and non-coding"` sequence.
● And later it assigned `color{Violet}"different regions in the sequence"` with functions (a term referred to as `color{Violet}"Sequence Annotation"`).
● One approach focused on identifying all the `color{Violet}"genes"` that expressed as `color{Violet}"RNA"` (referred to as `color{Violet}"Expressed Sequence Tags"` (`color{Violet}"ESTs"`).
● The other took the `color{Violet}"blind approach"` of simply sequencing the `color{Violet}"whole set of genome"` that contained all the `color{Violet}"coding and non-coding"` sequence.
● And later it assigned `color{Violet}"different regions in the sequence"` with functions (a term referred to as `color{Violet}"Sequence Annotation"`).