`->` To start the synthesis of new chain, special type of R.N.A. is required which is termed as R.N.A. Primer. The formation of R.N.A. primer is catalysed by an enzyme - R.N.A. Polymerase (primase). Synthesis of RNA-primer takes place in `5' ->- 3'` direction. After the formation of new chain, this R.N.A. is removed. For the formation of new chain Nucleotides are obtained from Nuceloplasm. In the nucleoplasm, Nucleotides are present in the form of triphosphaies like dATP, dGTP, dCTP, dTn) etc.
`->` During replication, the `2` phosphate groups of all Nucleotides are separated. In this process energy is yeilded which is consumed in D.N.A. replication. So, it is clear that D.N.A. does not depend on mitochondria for it's energy requirements.
`->` Energetically replication is a very expensive process .. Daoxyribonucleoside triphosphase serve duas purposes in addition to acting as substrates they provide energy for polymerisation.
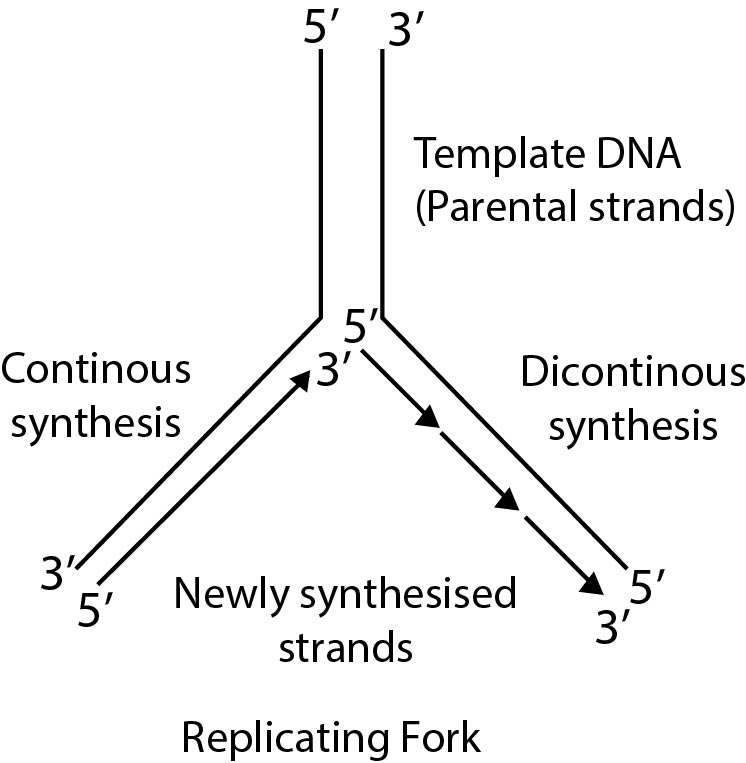
`->` The formation of new chain always takes place in 5' - 3' direction. 1\s a result of this, one chain of D.N.A. is continuously formed and it is termed as Leading strand. The formation of second chain begins from the centre and not from the terminal points, so this chain is discontinuous and is made up of small segments called Okazaki Fragments. This discontinuous chain is termed as Lagging strand. Ultimately all these segments joined together and a complete new chain is formed.
`->` The Okazaki segments are joined together by an enzyme DNA LiBase.(Khorana).
`->` The formation of new chains is catalysed by an enzyme DNA Polymerase. In prokaryotes it is of 3 types.
(1) DNA- Polymerase I :-This was discovered by KORNBERG (1957). So it is also called as 'kornberg's enzyme'. Kornberg also synthesized DNA first of all, in the laboratory. This enzyme functions as exonuclease. It separates RNA - primer from DNA and also fills the gap.lt is also known as DNA-repair enzyme.
(2) DNA - Polymerase II :- It is least reactive in replication process. It is also helpful in DNA-repairing in absence of DNA-polymerase-! and DNA polymerase-III
(3) DNA - Polymerase Ill :- This is the main enzyme in DNA - Replication. It is most important , It was discovered by Delucia and Cairns. The larger chains are formed by this enzyme. This is also known as Replicase. DNA -polymerase III is a complex enzyme composed of seven polypeptides
i.e. `alpha , sigma , theta , beta , gamma_1 , delta , gamma_2`
In Eucaryotes, there occur five types of DNA-polymerase enzyme.
(1) `alpha`-DNA - polymerase = 11 is concern with lagging strand synthesis.
(2) `beta`-DNA - polymerase = It concerned with DNA repair.
(3) `gamma`-DNA - polymerase = It concerned with replication of cytoplasmic DNA.
(4) `delta` -DNA - polymerase l1 is concern with leading strand synthesis.
(5) `sigma`- DNA polymerase = It is concern with proof reading.
`->` Thus DNA - Replication process is completed with the effect of different enzymes.
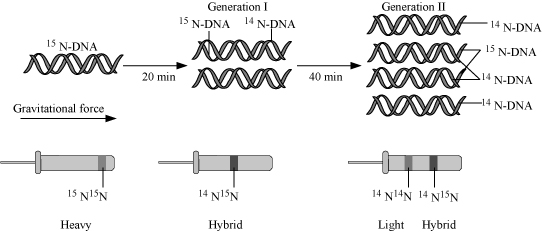
`->` In the semi conservative mode of replication each daughter DNA molecule receives one chain of polynucleotides
from the mother DNA - molecule and the second chain is synthesized.
`->` To start the synthesis of new chain, special type of R.N.A. is required which is termed as R.N.A. Primer. The formation of R.N.A. primer is catalysed by an enzyme - R.N.A. Polymerase (primase). Synthesis of RNA-primer takes place in `5' ->- 3'` direction. After the formation of new chain, this R.N.A. is removed. For the formation of new chain Nucleotides are obtained from Nuceloplasm. In the nucleoplasm, Nucleotides are present in the form of triphosphaies like dATP, dGTP, dCTP, dTn) etc.
`->` During replication, the `2` phosphate groups of all Nucleotides are separated. In this process energy is yeilded which is consumed in D.N.A. replication. So, it is clear that D.N.A. does not depend on mitochondria for it's energy requirements.
`->` Energetically replication is a very expensive process .. Daoxyribonucleoside triphosphase serve duas purposes in addition to acting as substrates they provide energy for polymerisation.
`->` The formation of new chain always takes place in 5' - 3' direction. 1\s a result of this, one chain of D.N.A. is continuously formed and it is termed as Leading strand. The formation of second chain begins from the centre and not from the terminal points, so this chain is discontinuous and is made up of small segments called Okazaki Fragments. This discontinuous chain is termed as Lagging strand. Ultimately all these segments joined together and a complete new chain is formed.
`->` The Okazaki segments are joined together by an enzyme DNA LiBase.(Khorana).
`->` The formation of new chains is catalysed by an enzyme DNA Polymerase. In prokaryotes it is of 3 types.
(1) DNA- Polymerase I :-This was discovered by KORNBERG (1957). So it is also called as 'kornberg's enzyme'. Kornberg also synthesized DNA first of all, in the laboratory. This enzyme functions as exonuclease. It separates RNA - primer from DNA and also fills the gap.lt is also known as DNA-repair enzyme.
(2) DNA - Polymerase II :- It is least reactive in replication process. It is also helpful in DNA-repairing in absence of DNA-polymerase-! and DNA polymerase-III
(3) DNA - Polymerase Ill :- This is the main enzyme in DNA - Replication. It is most important , It was discovered by Delucia and Cairns. The larger chains are formed by this enzyme. This is also known as Replicase. DNA -polymerase III is a complex enzyme composed of seven polypeptides
i.e. `alpha , sigma , theta , beta , gamma_1 , delta , gamma_2`
In Eucaryotes, there occur five types of DNA-polymerase enzyme.
(1) `alpha`-DNA - polymerase = 11 is concern with lagging strand synthesis.
(2) `beta`-DNA - polymerase = It concerned with DNA repair.
(3) `gamma`-DNA - polymerase = It concerned with replication of cytoplasmic DNA.
(4) `delta` -DNA - polymerase l1 is concern with leading strand synthesis.
(5) `sigma`- DNA polymerase = It is concern with proof reading.
`->` Thus DNA - Replication process is completed with the effect of different enzymes.
`->` In the semi conservative mode of replication each daughter DNA molecule receives one chain of polynucleotides
from the mother DNA - molecule and the second chain is synthesized.