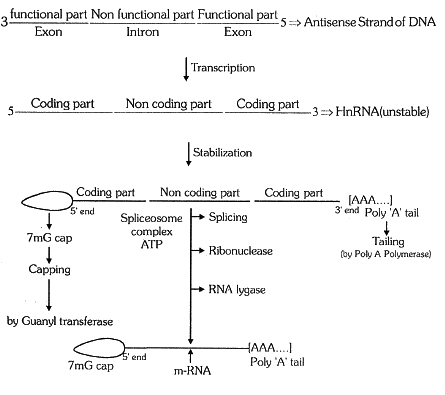
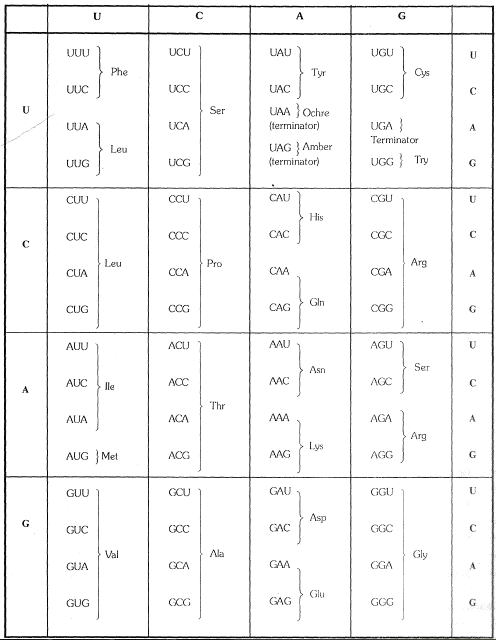
Transcription
`->` Formation of RNA over DNA template is called transcription. Out of two strand of DNA only one strand participates in transcription and called "Antisense strand".
`->` If both strands act as a template during transcription they would code for ENA molecule with different sequence and If they code for proteins the sequence of aminoacid in these protein would be different and another reason that if the two HNA molecule produced they would be complementary to each other and form a ds RNA which prevent translation of RNA.
`->` A gene is defined as the functional unit of inheritance. It is difficult to literally define a gene in terms of DNA sequence, because the DNA sequence coding for tENA or rENA molecule is also define a gene (But information of protein is present on the DNA segment which code mRNA. So generally it is
reffered for it)
`->` The functional unit of DNA involved in transcription is "Cistron " .
`->` RNA polymerase enzyme is involved in transcription. In eukaryotes there are three types of RNA polymerases.
# RNA polymerase-I for 28s rHNA, 18s rENA, 5.8s rENA synthesis.
# RNA polymerase-ll for m-HNA synthesis.
# RNA polymerase-Ill for t-rmA, 5s rENA, SnRNA synthesis.
`->` In eukaryotes RNA polymerase enzyme is made up of `10-15` polypeptide chains.
`->` Prokaryotes have only one type of ENA polymerase which synthesizes all types of HNAs.
`->` RNA polymerase of E. Coli has five polypeptide chains `beta , beta', alpha ., alpha`. and `sigma`.
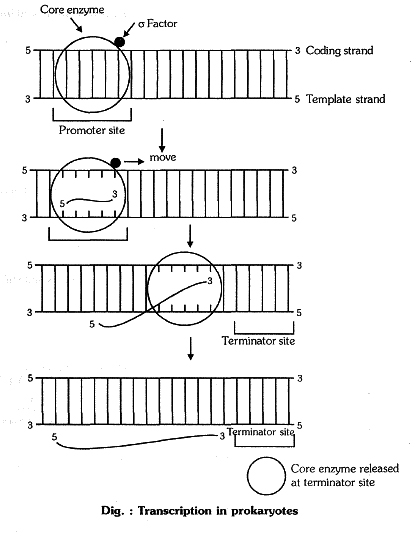
`->` `sigma` polypeptide chain is also known as a factor (sigma factor).
`->` Core enzyne + Sigma factor `=>` RNA Polymerase
(beta , beta' , alpha , alpha ) (sigma)
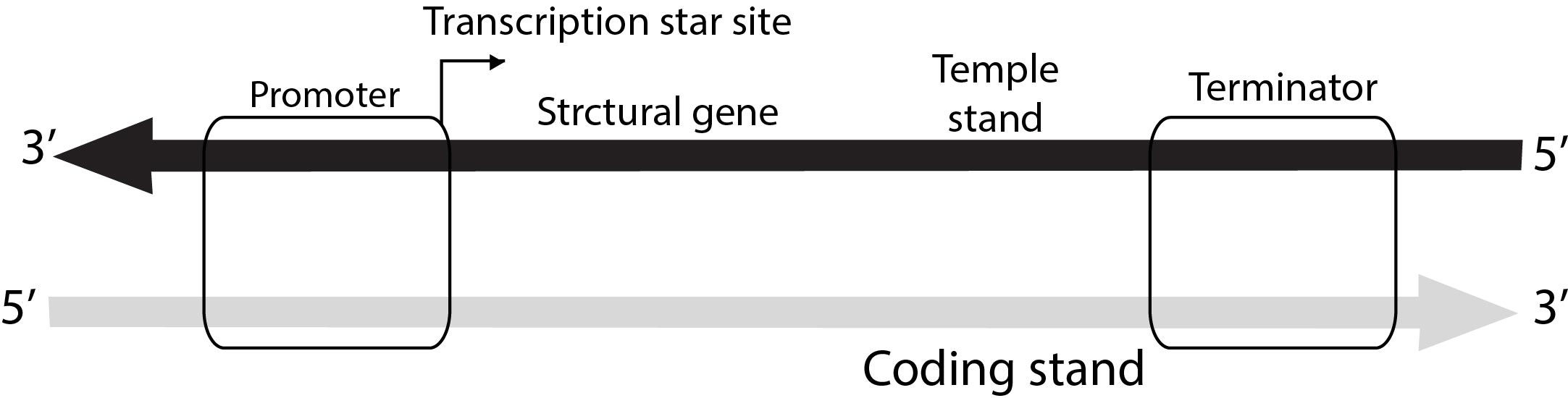
A transcription unit in DNA is defined primarily by the in three gigons in the DNA :-
(i) A promoter, (ii) The structural gene (iii) A terminator
Following steps are present in transcription -
`->` If both strands act as a template during transcription they would code for ENA molecule with different sequence and If they code for proteins the sequence of aminoacid in these protein would be different and another reason that if the two HNA molecule produced they would be complementary to each other and form a ds RNA which prevent translation of RNA.
`->` A gene is defined as the functional unit of inheritance. It is difficult to literally define a gene in terms of DNA sequence, because the DNA sequence coding for tENA or rENA molecule is also define a gene (But information of protein is present on the DNA segment which code mRNA. So generally it is
reffered for it)
`->` The functional unit of DNA involved in transcription is "Cistron " .
`->` RNA polymerase enzyme is involved in transcription. In eukaryotes there are three types of RNA polymerases.
# RNA polymerase-I for 28s rHNA, 18s rENA, 5.8s rENA synthesis.
# RNA polymerase-ll for m-HNA synthesis.
# RNA polymerase-Ill for t-rmA, 5s rENA, SnRNA synthesis.
`->` In eukaryotes RNA polymerase enzyme is made up of `10-15` polypeptide chains.
`->` Prokaryotes have only one type of ENA polymerase which synthesizes all types of HNAs.
`->` RNA polymerase of E. Coli has five polypeptide chains `beta , beta', alpha ., alpha`. and `sigma`.
`->` `sigma` polypeptide chain is also known as a factor (sigma factor).
`->` Core enzyne + Sigma factor `=>` RNA Polymerase
(beta , beta' , alpha , alpha ) (sigma)
A transcription unit in DNA is defined primarily by the in three gigons in the DNA :-
(i) A promoter, (ii) The structural gene (iii) A terminator
Following steps are present in transcription -