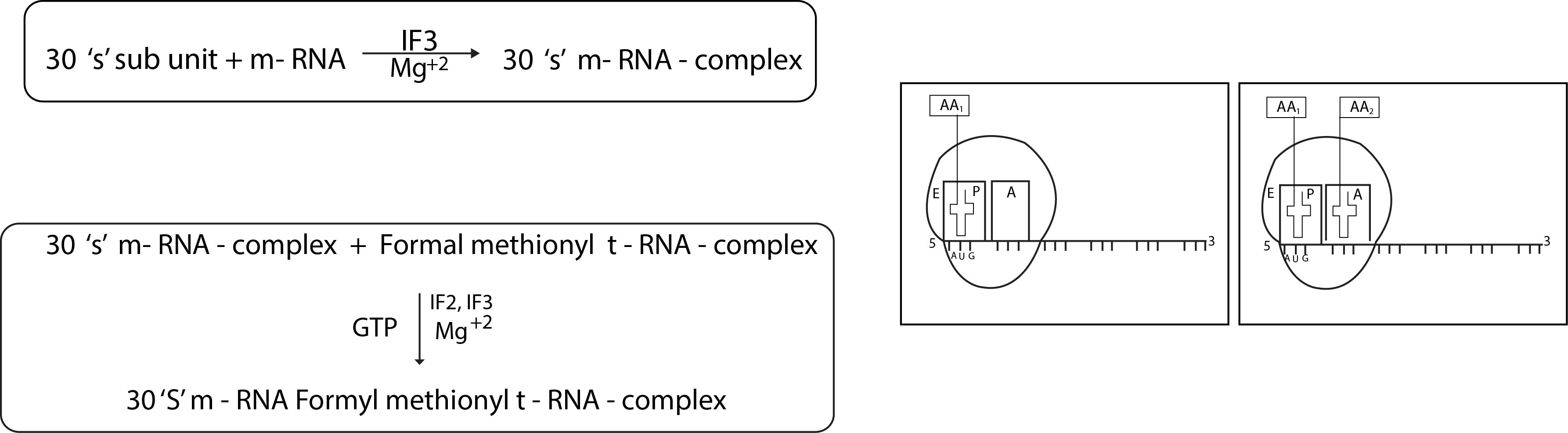
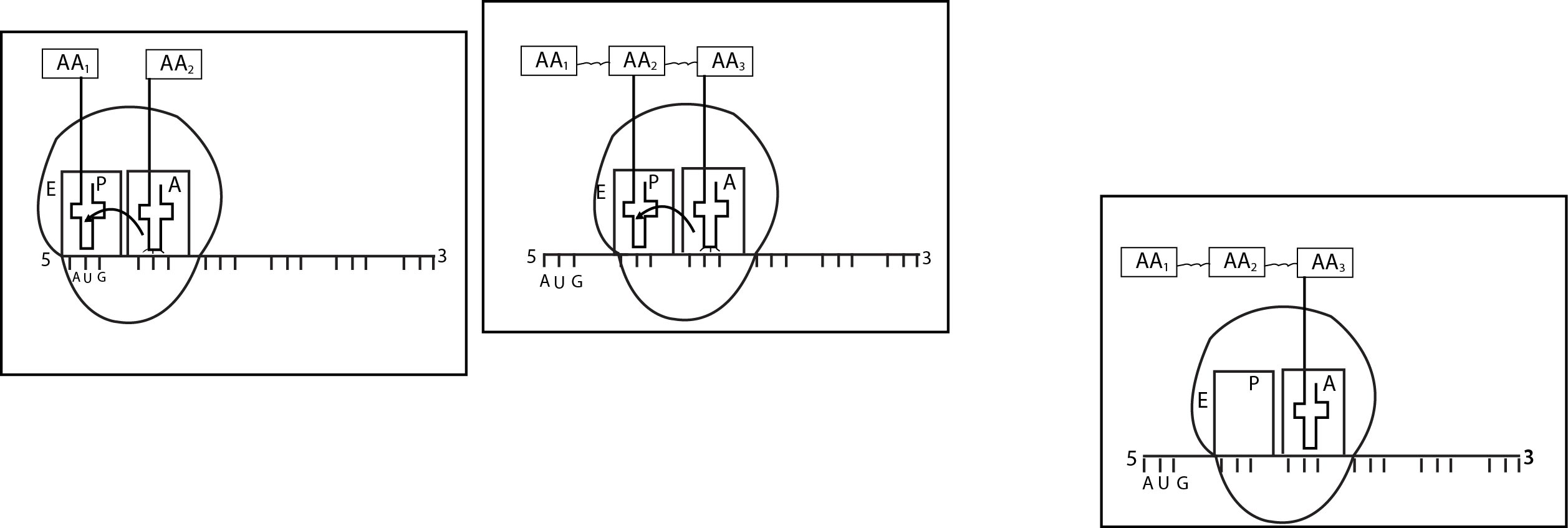
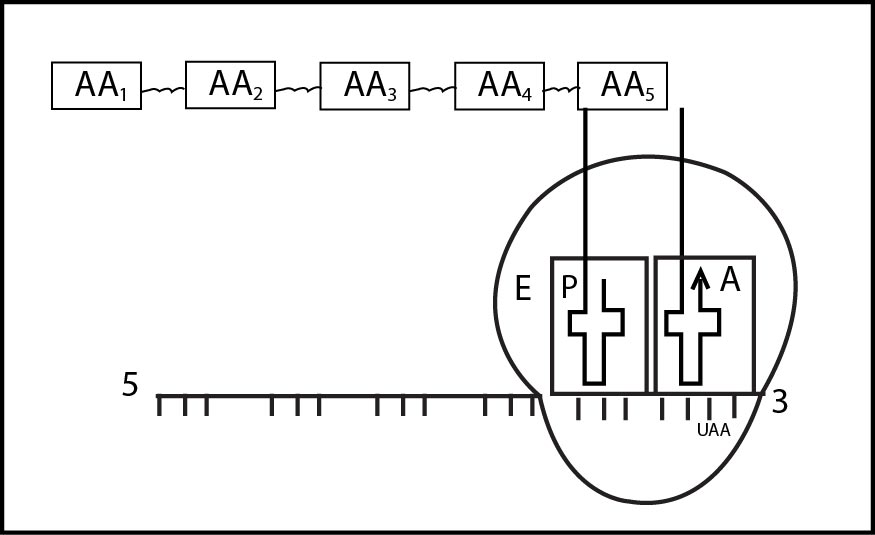
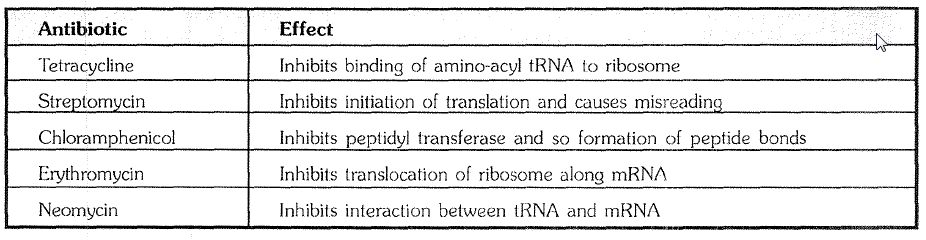
(1) Activation of Amino acid : -

`->` 20 types of amino acids participate in protein synthesis .
`->` Amino acid reacts with ATP to form "Amino acyl AMP enzyme complex" , which is also known as Activated
Amino acid'.
`->` This reaction is catalyzed by a specific 'Amino acyl t-RNA synthetase' enzyme .
`->` There is a separate 'Amino acyl t-RNA synthetase' enzyme for each kind of amino acid.
`->` Amino acid reacts with ATP to form "Amino acyl AMP enzyme complex" , which is also known as Activated
Amino acid'.
`->` This reaction is catalyzed by a specific 'Amino acyl t-RNA synthetase' enzyme .
`->` There is a separate 'Amino acyl t-RNA synthetase' enzyme for each kind of amino acid.