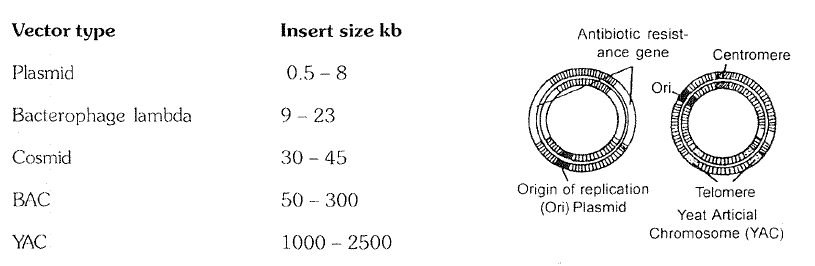
2. Vehicle DNA or Vector DNA.
The DNA used as a carrier for transferring a fragment of foreign DNA into a suitable host is called vehicle or vector DNA.
You know that plasmids and bacteriophages have the ability to replicate within bacterial cells independent of the control of chromosomal DNA.
The following are the features that are required to facilitate clonin~3 into a vector.
(i) Origin of replication (ori) : This is a sequence from where replication starts and any piece of DNA when linked to this sequence
can be made to replicate within the host cells. This sequence is also responsible for controlling the copy number of the linked DN/\. So.
if one wants to recover many copies of the target DNA it should be cloned in a vector whose origin support high copy number.
(ii) Selectable marker : In addition to 'ori', the vector requires a selectable marker. Normally, the genes encodin~J resistance to antibiotics such as ampicilin, chloramphenicol, tetracycline or kanamycin, etc., are considered useful selectable markers for E. coli.
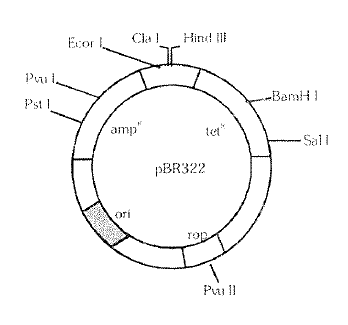
(iii) Cloning sites : In order to link the alien DNA, the vector needs, recognition sites for the commonly used restriction enzymes. The ligation of alien DNA is carried out at a restriction site present in one of the two antibiotic resistance genes. For example, you can ligate a foreign DNA at the Barn H site of tetracycline resistance gene in the vecter pBR322. The recombinant plasm ids will loss tetracycline resistance due to insertion of foreign DNA (insertional inactivation) now. it can be selected out from non-recombinant ones by plating the transformants on ampicillin containing medium. The transformants (plasmid transfer) growing on ampicillin containing medium are then transferred on a medium containing tetracycline. The recombinants will grow in ampicillin containin~J medium but not on that containin tetracycline. But non· recombinants will grow on the medium containing both the antibiotics. In this case. one antibiotic resistance gene helps in selecting the transforrnants.
Selection of recombinants due to inactivation of antibiotics is a cumbersome (troublesome) nu,rom because it requires simultaneous plating on two plated having different antibiotics. Therefore , alternative selectable markers have been developed which differentiate recombinants from non-recombinants on the basis of their ability to prcxluce colour in the presence of a chromogenic substrate. In this. a recombinant DNA is inserted within the coding sequence of an enzyme, which is referred to as insertional inactivation.
The presence of a chromogenic substrate X-gal (5-·bromo-4chloro-`beta`- D galacto pyranoside) gives blue coloured colonies if the plasmid in the bacteria does not have an insert. Presence of insert results into insertional inactivation of the [3-galactosidase (reporter enzyme) and the colonies do not produce any colour, these are identified as recombinant colonies.
`->` Vectors for cloning genes in plants and animals : A~3robactcrium tumifacicns, a pathogen of several dicot plants deliver a piece of DNA known as T-DNA' to transform normal plant cells into a tumour. Similarly, retroviruses in animals have the ability to transform normal cells into cancerous cells. A better understanding of the art of delivering ~1enes by pathogens in their eucaryotic hosts has generated knowledge to transform these tools of pathogens into useful vectors for delivering qenes of interest to humans. The tumour inducing (Ti) plasmid of Agrobacteriurn tumifaciens has now been modified into cloning vector which is no more pathogenic to the plants but is still able to use the mechanisms to (disarmed) and are now used to deliver desirdble genes into dninml cells. So, once a gene or a DNA fragment has been ligated into a suitable vector it is transferred into a bacterial, plant or animal host (where it multiplies).
You know that plasmids and bacteriophages have the ability to replicate within bacterial cells independent of the control of chromosomal DNA.
The following are the features that are required to facilitate clonin~3 into a vector.
(i) Origin of replication (ori) : This is a sequence from where replication starts and any piece of DNA when linked to this sequence
can be made to replicate within the host cells. This sequence is also responsible for controlling the copy number of the linked DN/\. So.
if one wants to recover many copies of the target DNA it should be cloned in a vector whose origin support high copy number.
(ii) Selectable marker : In addition to 'ori', the vector requires a selectable marker. Normally, the genes encodin~J resistance to antibiotics such as ampicilin, chloramphenicol, tetracycline or kanamycin, etc., are considered useful selectable markers for E. coli.
(iii) Cloning sites : In order to link the alien DNA, the vector needs, recognition sites for the commonly used restriction enzymes. The ligation of alien DNA is carried out at a restriction site present in one of the two antibiotic resistance genes. For example, you can ligate a foreign DNA at the Barn H site of tetracycline resistance gene in the vecter pBR322. The recombinant plasm ids will loss tetracycline resistance due to insertion of foreign DNA (insertional inactivation) now. it can be selected out from non-recombinant ones by plating the transformants on ampicillin containing medium. The transformants (plasmid transfer) growing on ampicillin containing medium are then transferred on a medium containing tetracycline. The recombinants will grow in ampicillin containin~J medium but not on that containin tetracycline. But non· recombinants will grow on the medium containing both the antibiotics. In this case. one antibiotic resistance gene helps in selecting the transforrnants.
Selection of recombinants due to inactivation of antibiotics is a cumbersome (troublesome) nu,rom because it requires simultaneous plating on two plated having different antibiotics. Therefore , alternative selectable markers have been developed which differentiate recombinants from non-recombinants on the basis of their ability to prcxluce colour in the presence of a chromogenic substrate. In this. a recombinant DNA is inserted within the coding sequence of an enzyme, which is referred to as insertional inactivation.
The presence of a chromogenic substrate X-gal (5-·bromo-4chloro-`beta`- D galacto pyranoside) gives blue coloured colonies if the plasmid in the bacteria does not have an insert. Presence of insert results into insertional inactivation of the [3-galactosidase (reporter enzyme) and the colonies do not produce any colour, these are identified as recombinant colonies.
`->` Vectors for cloning genes in plants and animals : A~3robactcrium tumifacicns, a pathogen of several dicot plants deliver a piece of DNA known as T-DNA' to transform normal plant cells into a tumour. Similarly, retroviruses in animals have the ability to transform normal cells into cancerous cells. A better understanding of the art of delivering ~1enes by pathogens in their eucaryotic hosts has generated knowledge to transform these tools of pathogens into useful vectors for delivering qenes of interest to humans. The tumour inducing (Ti) plasmid of Agrobacteriurn tumifaciens has now been modified into cloning vector which is no more pathogenic to the plants but is still able to use the mechanisms to (disarmed) and are now used to deliver desirdble genes into dninml cells. So, once a gene or a DNA fragment has been ligated into a suitable vector it is transferred into a bacterial, plant or animal host (where it multiplies).